Troubleshooting in R
Every one who uses R, beginner or advanced, faces errors in their code. In fact, learning how to read errors messages and troubleshoot them is one of the most important aspects of learning R. The first half of this page goes over some good strategies to troubleshoot and will give you a troubleshooting challenge. The second half of the page lists some of the most common mistakes people do. You can refer back to this page whenever you face a problem to help you troubleshoot it.
1 - Common strategies to troubleshoot
1.1. Keep calm and read the error message
The first thing to do when your code is not working is to read the error message. As frustrating as it can be to see an error message, they are in fact there to help you identify what the mistake was. For example, let’s look at the error message below:
# Create an object with values
values <- c(1, 2, 3, 4, 5)
# Calculate the mean of these values
mean(value)
## Error: object 'value' not found
Instead of outputting the mean value, R is saying that the object value was not found. This might create confusion at first, as you would think you just created the object in the line above. But if R is not finding the object, you more likely did not create it. A close inspection shows that you created the object values, not value. This is a simple typo that can be easily fixed:
# Create an object with values
values <- c(1, 2, 3, 4, 5)
# Calculate the mean of these values
mean(values)
## [1] 3
Sometimes errors messages are more complicated than this, for example:
# Load package
library(tidyverse)
# Create vector of values
values <- c(1, 1, 2, 2, 3, 3)
# Identify unique set of distinct values
distinct(values)
## Error in UseMethod("distinct"): no applicable method for 'distinct' applied to an object of class "c('double', 'numeric')"
Here, the error message might seem a bit daunting, but if you read with care, you can identify that R is telling you that it can not use the function distinct to an object of the type double or numeric (which your vector values is). This might prompt you to open the help of the function (see section below), where you will see that the distinct() function takes a data.frame as its first argument, and not a vector. This is why this code is failing. You might then remember that the function unique() is the one you wanted to use, which does the same but for vectors:
# Load package
library(tidyverse)
# Create vector of values
values <- c(1, 1, 2, 2, 3, 3)
# Identify unique set of distinct values
unique(values)
## [1] 1 2 3
There are numerous potential error messages and the point there is not to go through them all, but to show you that errors messages can be very helpful to identify what the error is.
1.2. Double-check your code
After reading the error message and identifying what the mistake likely is, double-check your code with attention. Most errors are usually caused by simple typos or forgotten closing brackets in the code (see list of common mistakes below).
1.3. Read function and package help
If you are facing an error when using a particular function, it is always helpful to open the documentation for that function. To do that, you can run help(function_name) or ?function_name in the console. Depending on the configurations of your RStudio, you can also open the help documentation by typing the function name (in the code editor or console), and then hitting the button F1.
Once you do that, the documentation for the function will open on the bottom right panel of your RStudio Interface, as long as you have loaded the package that the contains the function.
For example, let’s try opening the help for the distinct() function that we attempted to use above:
?distinct
The help page will contain all or some of the following sections:
- Description: a brief description of what the function does
- Usage: the code to use the function, showing arguments and their default values
- Arguments: a detailed description of each argument, including what type of object is expected
- Details: any important details about the function
- Value: a description of the object returned by the function
- Methods: specific methods for different object types, if applicable
- See Also: related functions that might be useful
- Examples: examples of how to use the function
In the distinct() function help, you can see under Arguments that the argument .data expects “A data frame, data frame extension (e.g. a tibble), or a lazy data frame (e.g. from dbplyr or dtplyr).”. That is why we could not use the function on a vector.
Besides function documentation, R packages also contain useful documentation. Many packages have vignettes, which are extended explanation of how to use the package, often including examples of how to use the functions in the package. You can assess the vignette by searching for the package webpage or typing vignette("vignette-name") in the console.
1.4. Run one line at a time
We often run multiple lines of code together to perform an operation, but when an error message appear, we might not know in which line the error is. It it thus good practice to run each line at a time to find an error.
For example, this code results in an error:
# This loads the built-in dataframe diamonds
data(diamonds)
# Create a derived column: price per carat
diamonds$price_per_carat <- diamonds$price / diamonds$carat
# Filter to a single cut
ideal <- subset(diamonds, cut == "Ideal")
# Filter to one type of clarity
ideal_vs2 <- subset(ideal, claritty == "VS2")
## Error in eval(e, x, parent.frame()): object 'claritty' not found
# Get the mean price per carat of the subset
mean_ppc <- mean(ideal_vs2$price_per_carat)
## Error: object 'ideal_vs2' not found
# check result
mean_ppc
## Error: object 'mean_ppc' not found
The error message already suggest that the error was where we calling clarity, but lets run each line of code to check:
data(diamonds)
This executes correctly, that is, the object is correclt loaded!
# This loads the built-in dataframe diamonds
data(diamonds)
# Create a derived column: price per carat
diamonds$price_per_carat <- diamonds$price / diamonds$carat
The calculation of price per carat also seems to be working
# This loads the built-in dataframe diamonds
data(diamonds)
# Create a derived column: price per carat
diamonds$price_per_carat <- diamonds$price / diamonds$carat
# Filter to a single cut
ideal <- subset(diamonds, cut == "Ideal")
The first subset also seems to work.
# This loads the built-in dataframe diamonds
data(diamonds)
# Create a derived column: price per carat
diamonds$price_per_carat <- diamonds$price / diamonds$carat
# Filter to a single cut
ideal <- subset(diamonds, cut == "Ideal")
# Filter to one type of clarity
ideal_vs2 <- subset(ideal, claritty == "VS2")
## Error in eval(e, x, parent.frame()): object 'claritty' not found
A-ha, there seems to be an error here! Once you know where the errors is, you can then more easily troubleshoot it. In this case, the problem is that there is a type in the name of the variable, it should be clarity not claritty:
# This loads the built-in dataframe diamonds
data(diamonds)
# Create a derived column: price per carat
diamonds$price_per_carat <- diamonds$price / diamonds$carat
# Filter to a single cut
ideal <- subset(diamonds, cut == "Ideal")
# Filter to one type of clarity
ideal_vs2 <- subset(ideal, clarity == "VS2") # Fixed typo
# Get the mean price per carat of the subset
mean_ppc <- mean(ideal_vs2$price_per_carat)
# check result
mean_ppc
## [1] 3814.12
1.5. Restart R session
When we are actively writing code, it is common to test out possibilities, which often require creating test objects, reordering lines of code, and loading new packages. In this process, sometimes we accidentally overwrite an object and load a package that masks a function we are using. An easy to make sure you didn’t do this is to clear your working environment of all objects, restart your R session, and then rerun the entire script.
To clear the working environment, you can run rm(list =ls()) in your console, or click the “Clear objects from workspace” button in the top right. This will remove all objects.
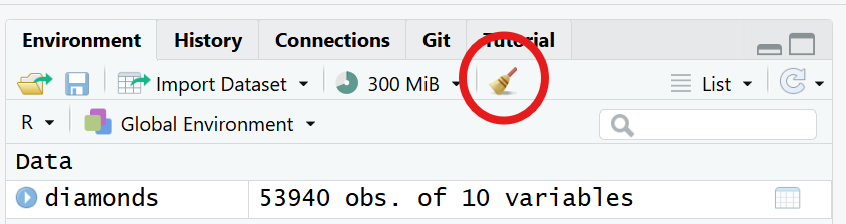
To restart your R session, click on Session at the top, and then on Restart R. This will remove any loaded packages. When rerunning your script again, R will load only the packages that you wrote in your script, so make sure your script loads any packages you need.
1.6. Look for similar errors online
Chances are, someone already ran into an error similar to yours. A quick online search often redirects you to a forum such as StackOverflow where someone already posted a question similar to yours and probably got an answer. When doing this online search, it is often helpful to search using specific terminology such as: the action you want to perform, the language you are using, and the specific style/technique you are using. For example, you could search: how to rename a column (i.e. the action) in R (i.e. the language) using base R (i.e. the style/technique of coding).
1.7. Take a break
Sometimes, the best way to troubleshoot an issue in R is just to take a break and come back later with a fresh mindset. Errors can be frustrating and the frustation and anger can prevent you from finding simple mistakes. Taking a break and coming back later usually works to find those mistakes.
1.8. Ask someone for help
As has been said, the best way to learn is to try to troubleshoot the error by yourself. However, if that does not work, you can always ask for help, both in forums in the internet, or to someone you know who might be a more advanced R user. If you are posting on a particular forum such as StackOverflow, don’t forget to read their guidelines about how to post a question.
1.9. A note about using AI
You might have noticed that, up until now, there was no mention of AI. That is because we encourage you to try to troubleshoot your errors by yourself, which is the best way to learn how to use R. However, AI can also be used adequately to help you troubleshoot, as long as you are not using AI as a way to avoid learning what the error was. For example, instead of pasting your problematic code and asking AI to fix it without understanding what went wrong, you could ask AI what an error message is referring to, or ask it to find typos in your code without editing your code.
Moreover, remember that AI models can also hallucinate and produce incorrect code. You don’t want to find yourself in a situation like the reddit user below, who didn’t check their code and ended up to get their entire home directory wiped out…
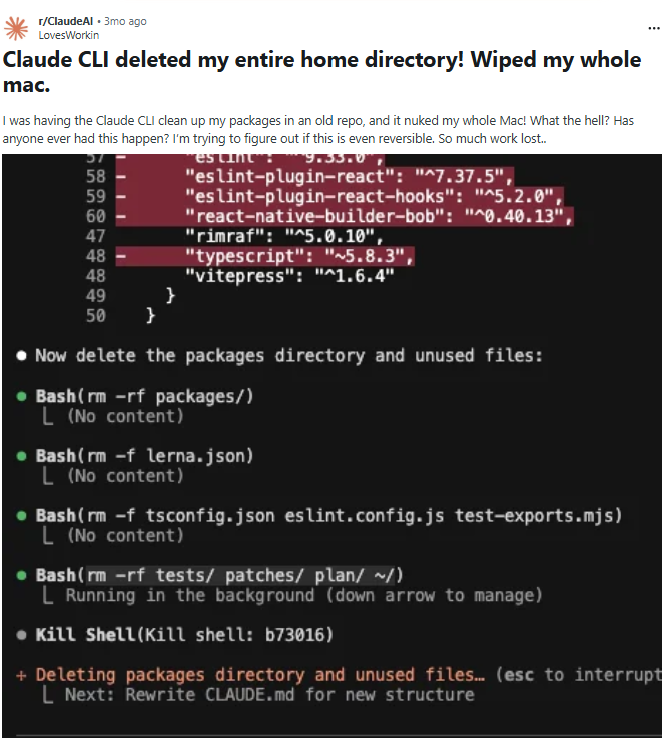
The lesson here is that you need to understand enough about R to be able to evaluate their output. If AI is doing all your coding for you and you don’t learn the underlying logic and syntax of R, it will be harder for you to verify their output. Remember, you should think of AI as your research assistant, and you are still responsible for any code produced by AI. That being said, AI can definitely be a helpful tool, especially if you use it with the right attitude and know what and how to ask for the most useful responses. While this workshop doesn’t cover best practices for using AI in coding, you can find some useful tips here and here.
1.10 Troubleshooting challenge
⭐ Troubleshoot the code below
Now that we have gone over some of the best strategies to troubleshoot your code, see if you can find and correct the errors in the code below. If you want, you can also scroll down the Common mistakes section below for a list of the most common errors you might encounter.
# This is not part of the challenge, but please run this line of code
# in your console to start a new and clean R Session
.rs.restartR()
# Load the tidyverse package (which contains the diamonds dataset)
library(tidyver)
# Load dataset
data(diamonds)
# View the first rows of the dataset
head(diamond)
# View the column names
namesdiamonds)
# Get the mean price of diamonds
mean(diamonds$Price)
# Calculate price per carat
diamonds$price / diamonds$carat
# Get mean price per carat
mean(diamond$price_per_carat)
# filter the dataset for the diamonds with Ideal cut quality
subset(diamonds cut == "Ideal")
# Get the average price per carat of diamonds with Ideal cut quality
mean(diamondsIdeal$price_per_carat)
View the corrected code
# Load the tidyverse package (which contains the diamonds dataset)
library(tidyverse) # correct spelling for the tidyverse package
# Load dataset
data(diamonds)
# View the first rows of the dataset
head(diamonds) # correct spelling for the diamonds objects
# View the column names
names(diamonds) # added a missing parenthesis
# Get the mean price of diamonds
mean(diamonds$price) # corrected capitalization for the column price
# Calculate price per carat
diamonds$price_per_carat <- diamonds$price / diamonds$carat # assigned the result to a new column in the dataset
# Get mean price per carat
mean(diamonds$price_per_carat) # corrected spelling in dataset name
# filter the dataset for the diamonds with Ideal cut quality
diamondsIdeal <- subset(diamonds, cut == "Ideal") ## added missing comma, and saved in a new object
# Get the average price per carat of diamonds with Ideal cut quality
mean(diamondsIdeal$price_per_carat) # nothing to correct
2 - Common mistakes
Here is a list of common mistakes all R users do. If you are facing a problem in your code, going through these is probably a good start to try to figure out what can be happening in your code.
2.1. Capitalization
You typed a lower-case when it should have been an upper-case, or vice-versa. R is case-sensitive so this will result in an error for functions and objects. For example, calling Mean() won’t run and result in an error code as the name of the function is mean(), all lower caps. Or you named the object MyData when you first created it but then later you are calling the object mydata.
2.2. Misspelling
You misspelled a function name or an object name. R Studio does not highlight spelling errors in code like in Microsoft Word, so you have to pay attention to this when typing. For functions and arguments that have both British and American spelling (e.g. colour/color), R accepts both versions.
2.3. Closing punctuation
Your forgot to close a parenthesis, curly bracy, square bracket, or quotation mark. You will know that this is happening because R will expect additional code to continue, so the console will show a + instead of >. You will also see a red X on the left side of the line in the code editor.
Alternatively, instead of not including a closing punctuation, you might have added it to the wrong location. The code might even run, but the results will be weird, or an error message will appear. For example, to get the average of a list of numbers going from 1 to 100 in steps of 5 you would write mean(seq(1, 100, by = 5)). However, if you write: mean(seq(1, 100), by = 5) (note the closing parenthesis before the by argument instead of after), the code will run, but the result is wrong.
2.4. Continuing punctuation
You forgot to add a comma before moving on to the next indented line. Indenting lines makes the code easier to read, but it can be easy to forget a comma or other punctuation. For example:
# Average of a list of numbers going from 1 to 100
mean(seq(1, 100 # missing comma
by = 5))
## Error in parse(text = input): <text>:3:10: unexpected symbol
## 2: mean(seq(1, 100 # missing comma
## 3: by
## ^
# Average of a list of numbers going from 1 to 100
mean(seq(1, 100, # added comma
by = 5))
## [1] 48.5
If you are familiar with tidyverse (if not, check out the Data manipulation and visualization workshop, this error might also mean that you forgot to add a pipe, or left a pipe open. Ifa pipe is left open and there are no lines of code after, you will note this because the console will show a + instead of >. However, if you have other code after, R will continue to run the code and it will shown an error, for example:
# Calculate and get new variables
diamonds2 <- diamonds |>
mutate(price200 = price - 200,
price300 = price - 300) |>
select(cut, price200, price300) |> # forgotten open pipe
# another piece of code after
mean(diamonds2$price200)
## Warning in mean.default(select(mutate(diamonds, price200 = price - 200, :
## argument is not numeric or logical: returning NA
To correct:
# Calculate and get new variables
diamonds2 <- diamonds |>
mutate(price200 = price - 200,
price300 = price - 300) |>
select(cut, price200, price300) # removed pipe
# another piece of code after
mean(diamonds2$price200)
## [1] 3732.8
2.5. Overwritting objects
You accidentally overwrote an object that you needed. This might be written in the code editor, or you might have done that in your console without noticing it. A good way to check is to always preview the objects you are using to make sure they are what you want them to be. For example, you can use head() to preview the first rows of a dataframe to make sure they look like how they should, or even ask R what type of object it is with class().
2.6. Not assigning objects
You did not save an object that you wanted to save. This is very common at the beginning, when you are still not used to assigning data to objects using <- so that R can save the object in the workspace and use it later.
For example, imagine you had this piece of code
# Calculate a new variable about price
diamonds$price - 100
# another piece of code after
mean(diamonds$price100)
## [1] 226 226 227 234 235 236 236 237 237 238 239 240 242 244 245
## [16] 245 248 251 251 251 251 252 253 253 253 254 255 257 257 257
## [31] 302 302 302 302 302 302 302 302 303 303 303 303 303 303 303
## [46] 303 303 303 304 304 304 304 304 304 304 305 305 305 305 305
## [61] 452 452 452 452 452 453 453 453 453 453 453 454 454 454 454
## [76] 454 454 454 454 454 454 454 454 454 454 454 454 454 454 454
## [91] 2657 2657 2657 2659 2659 2659 2659 2659 2660 2660
## Warning: Unknown or uninitialised column: `price100`.
## Warning in mean.default(diamonds$price100): argument is not numeric or logical:
## returning NA
## [1] NA
Because you never saved the calculation of the new variable into the diamonds object, R cannot find it to calculate the mean. This is the corrected piece of code:
# Calculate new variable about price, and assign to the diamond object using <-
diamonds$price100 <- diamonds$price - 100
# another piece of code after, using the new object
mean(diamonds$price100)
## [1] 3832.8
2.7. Wrong working directory
You are using the wrong working directory. The working directory is the folder R is looking at to search for files you want to upload. However, if you forgot to specify the working directory (or specified a wrong one), R will not be able to find the files you are trying to upload. A quick way to check your working directory is to run getwd() in your console. If the directory is wrong, you can use setwd() to set the correct directory or click on Session > Set Working Directory > Choose Directory.
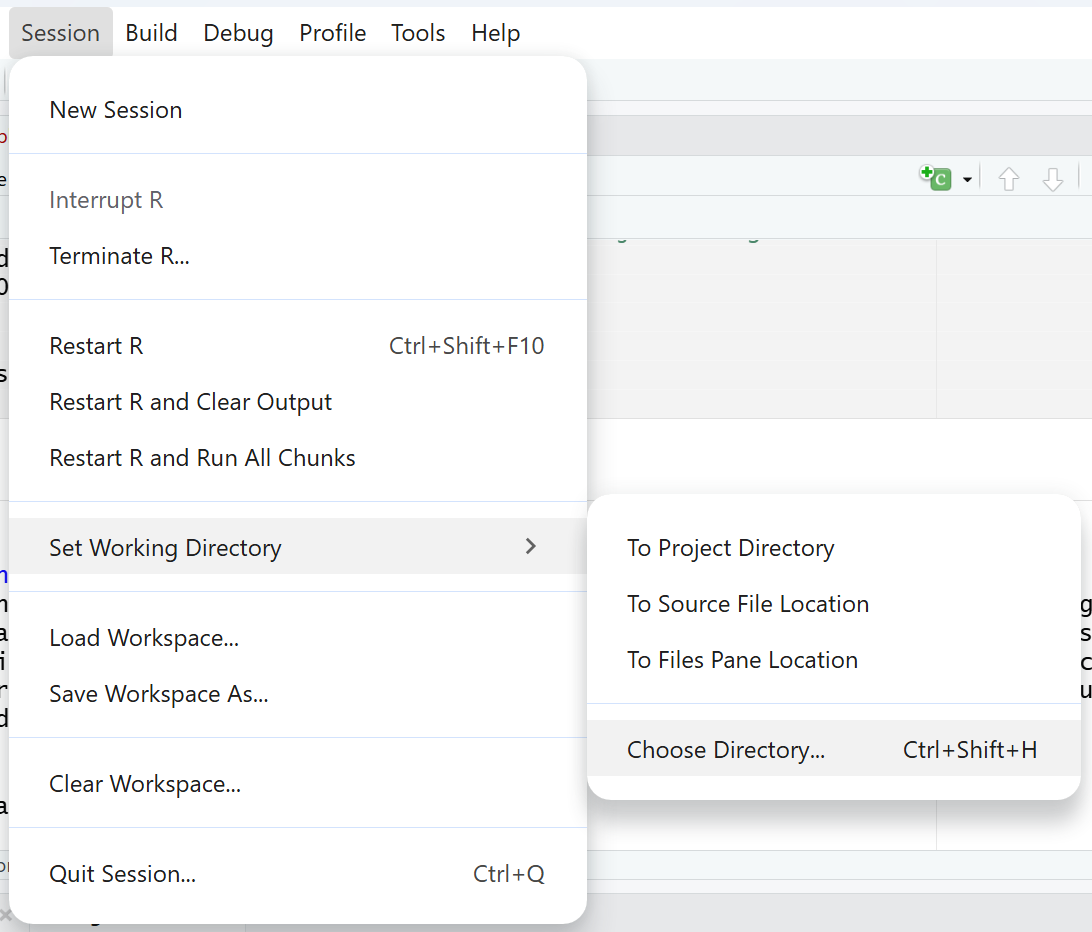
2.8. Package not loaded
The package you want is not loaded. It is important to make sure all the relevant packages are loaded. Whenever you close R, the packages are unloaded, so you need to load your packages every time you open R. A typical sign that this is an mistake is if the error message reads that a certain function could not be found.
2.9. Function masked by other packages
You are using a function that is masked by another a package. What does this mean? Sometimes, different packages have functions with the same name. When that happens, R will use the functions from the package that was more recently loaded. This is called “masking”. If you want to use the function from the other package, but R is using the function from the most recent package, the code will probably break due to different arguments. The error message will say something indicating that you have a wrong argument or your usage of the function is wrong.
When you suspect that, a good way to investigate further is to call the help of the function and check to which package it belongs too. Packages are shown on the top left of the help documentation, between {} after the function name. You can then see if the package is the one you intend to use.
Another way is to pay attention to messages that appear when loading the packages. If the loaded package is masking a function, you will see a message that says: “The following object is masked from ‘package:xxx : function_name’”. You then know to pay attention to masking when using those functions.
If you want to use the function from the other package, you need to specify that by using package::function(). For example, the tidyverse has a function called filter(), which masks a function of the same name from the base stats package. If you want to use the package from the stats package, you can specify: stats::filter().
2.10. Wrong package version
You package version does not work with your R version. From time to time, both base R and packages are updated. If you haven’t worked in your code for a while and have updated base R since, you might find that the version of the package that you have is not in sync with the version of R. Or sometimes the opposite happens, you downloaded a package that is for a newer version of R that you currently have. Sometimes this will not cause any issues, but it might cause some functions to stop working.
This is detected when loading a package. You will see a warning message that says: “package ‘package_name’ was built under R version X.X.X”. you can then run getRversion() in your console to check if your version of R is newer or older than the package.
If you need to update R, you can simply just re-install R from the CRAN website.
If you need to update the package, the easiest thing is to re-install the package using install.packages().